Key Result
This project is currently in progress, but it aims to contribute greatly to clubroot management and to empower producers to make informed decisions about the best canola cultivar to use based on P. brassicae diversity in each field.
Project Summary
Overview
Among the diseases affecting canola, clubroot (caused by the pathogen Plasmodiophora brassicae) is one of the biggest threats for the Canadian canola industry. One of the clubroot related challenges facing the industry is the need for reliable screening techniques to assess the durability of new forms of clubroot cultivar resistance and the identification and categorization of new and existing clubroot pathotypes.
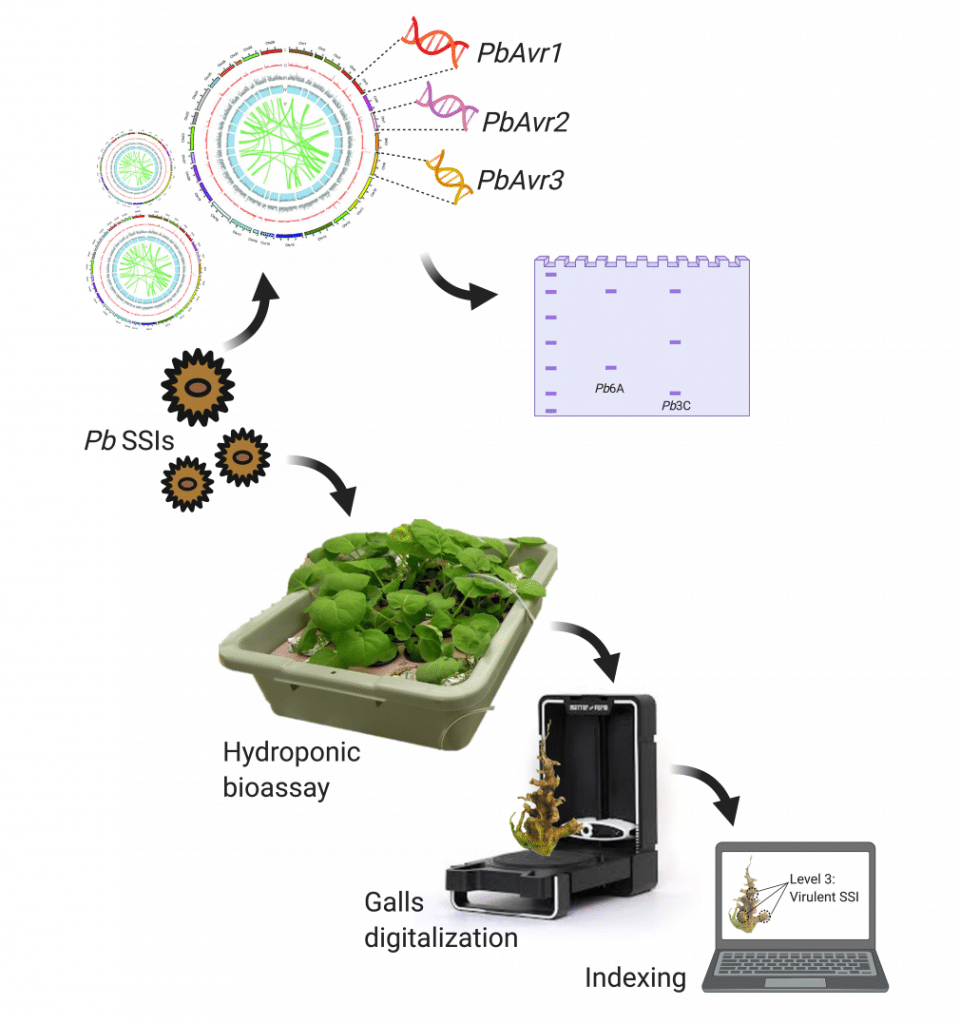
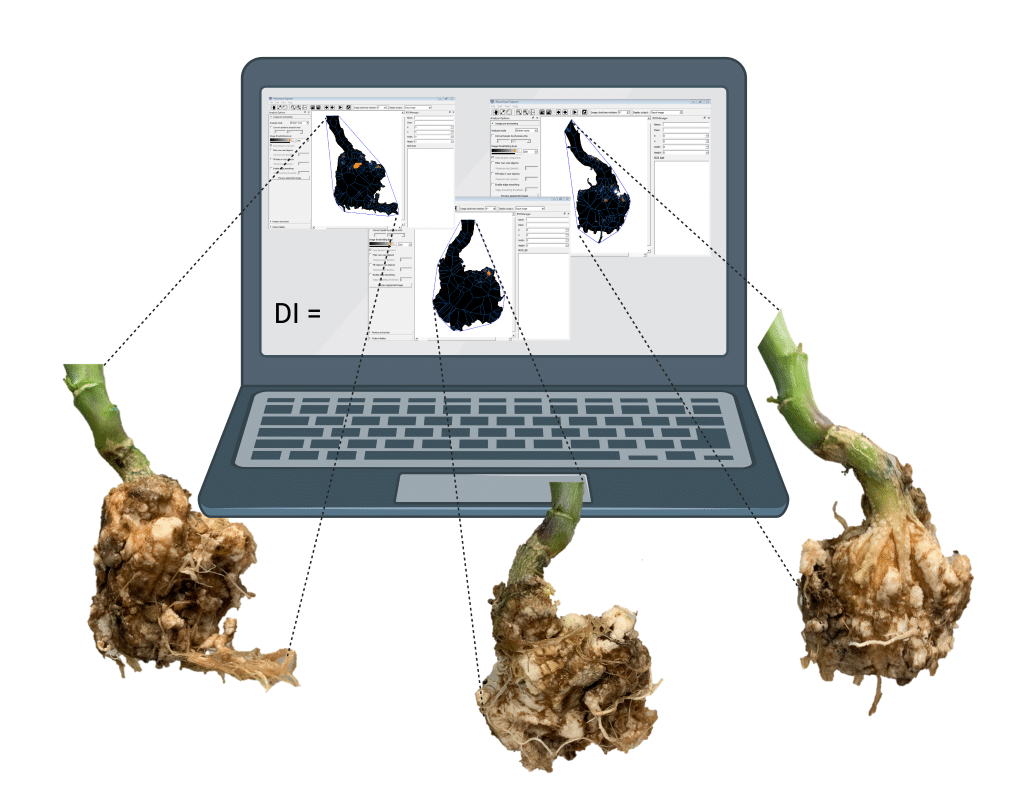
The proposed research addresses both of these important objectives by developing a bioassay to phenotype the interaction between canola and P. brassicae, identifying P. brassicae single spores isolates (SSI) avirulence markers, and developing a multiplex PCR assay able to differentiate P. brassicae isolates.
With precise knowledge of the diversity of pathotypes in the field and their virulence towards commercial canola varieties, it would be possible to tailor the selection of resistant canola varieties to use in an infested field for the longest possible time. For this purpose, it becomes essential to obtain:
(i) a fast and reliable phenotyping system
(ii) a wide and accurate genomic collection of P. brassicae pathotypes in Canada
(iii) a molecular assay that allows fast and precise identification of avirulent pathotypes.
Pérez-López’s research is precisely targeted to fill those existing gaps existing in the canola industry and will be essential for canola farmers, breeders, and the canola industry in general contributing to long-term management of the clubroot disease. Overall, this research will contribute greatly to P. brassicae management and to empower producers to make informed decisions about the best canola cultivar to use based on P. brassicae diversity in each field.
Objectives
- To optimize a hydroponic bioassay to phenotype the interaction between canola and P. brassicae.
- To sequence the genome of all SSIs and identify P. brassicae SSI avirulence markers.
- To design and implement a multiplex PCR assay able to differentiate P. brassicae isolates.
The schematic representation (above) depicts the proposed research (objective 1-3). Identification of Avirulent/Virulent phenotypes and identification of PbAvr genes that can be used as avirulence markers in order to develop a fast and accurate diagnostic test able to identify avirulent and virulent pathotypes in a field.
Progress
Perez-Lopez published the Profiling the clubroot pathogen, a cruciferous killer – Molecular Plant Pathology Highlight blog referencing his December 2022 publication: The clubroot pathogen Plasmodiophora brassicae: A profile update.