Key Result
This study found seven clubroot-resistance genes in B. rapa, B. nigra and B. oleracea and entered three new CR lines into canola co-op trials, big steps to improve the sustainability of canola production with increasing scope and intensity of clubroot across the Prairies.
Project Summary
Purpose
Clubroot was found in canola in new counties in western Canada in 2018, causing increased concern and likely increased interest in growing clubroot-resistant (CR) varieties. Annual disease surveys, research findings and lab samples continue to gather information on the distribution and prevalence of different pathotypes across the Prairies. However, a strong need to explore new resistant sources to stay ahead of the resilient and complex disease still remains.
This project aimed to discover new CR genes with novel traits from new B. rapa, B. oleracea and B. nigra sources, to develop markers for use in marker-assisted breeding and to investigate resistance mechanisms by different CR genes. It also involved the pyramiding of different resistance genes for the development or re-synthesizing of elite B. napus canola lines and of B. carinata canola germplasm (all of which involved many steps and required an enormous amount of careful, detailed work).
Results
The highlights of the genetic material created, developed or discovered throughout this project include:
• Genetic mapping of several B. rapa, B. nigra and B. oleracea lines have led to the discovery of seven CR genes. Gene-specific markers have also been developed for several of these CR genes.
• A CR B. carinata line (which came from a line with complete resistance to a broad spectrum of clubroot races) was created and is currently at the pre-commercialization stage.
Testing hundreds of lines in preliminary trials for yield and nursery plots for yield, agronomic and seed quality traits as well as in greenhouses or field plots for clubroot resistance resulted in 12 elite CR lines (that have been converted to cytoplasmic male-sterile lines for use as female parents in continued development of CR hybrids).
• Three canola cultivars carrying more than one CR genes have been entered in private co-op trials (and could be registration as early as 2019 if selected by seed companies) and two CR canola cultivars have been developed in collaboration with industry.
The key methods and analytical techniques developed throughout this project include:
• Leading-edge technologies (transcriptome, proteome, metabolome and synchrotron-based spectroscopy) have been developed to better study plant-pathogen interactions and resistance mechanisms, which helps optimize CR genes deployment for maximum efficacy and durability.
• Through an extensive process, it was determined that a new mapping method used in this study (mapping by sequencing) can successfully be used for genetic mapping.
• After multiple steps of analysis and sequencing, CR gene sequences and their expression could be used to distinguish the commonly used CR genes from novel CR genes.
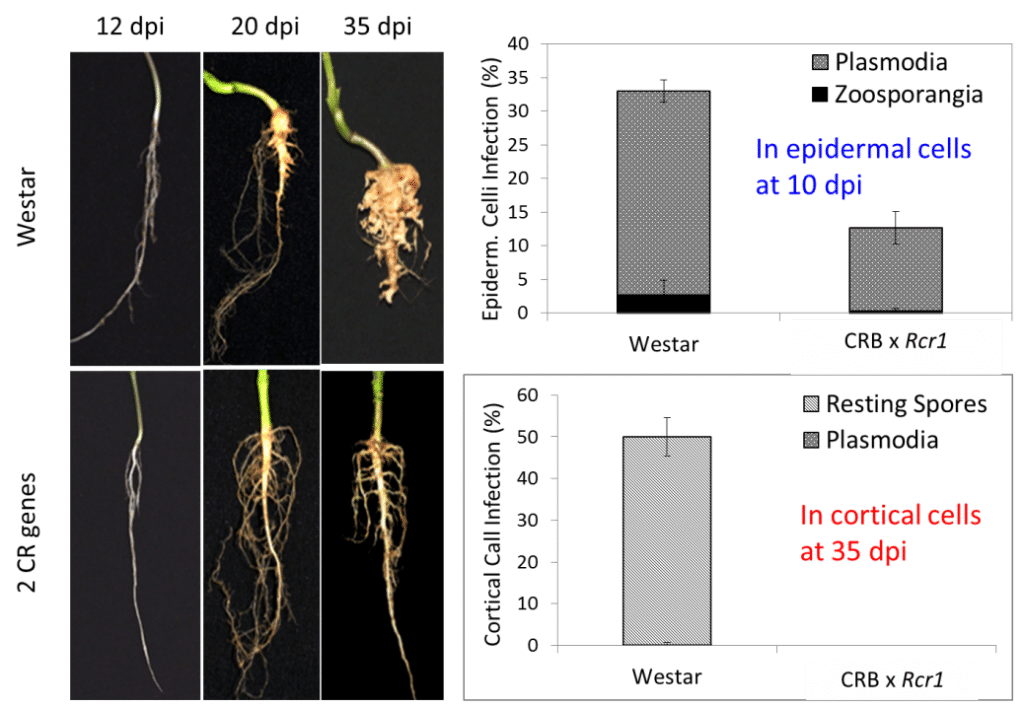
To better understand the mechanisms of the CR gene which affect the plant’s biochemistry and make it resistant, a synchrotron-based study was conducted to look at changes in cell-wall components associated with clubroot resistance.
The results showed an increased production of lignin and phenolics. Additional fluorescence microscopy on a moderately resistant canola hybrid carrying two CR genes confirmed the increased cell-wall lignification coinciding with reduced infection in root cortical tissues 20 to 35 days after inoculation with pathotype 5X of clubroot pathogen (see Figure 1). This is a very useful disease mechanism discovery.