Key Result
The diversity in the blackleg pathogen population shows the potential for resistance erosion, so continued monitoring is needed. To manage selection pressure/resistance breakdown, judicious uses of R genes is important for genetic control of blackleg. If an R gene is used repeatedly, there will be selection pressure for virulent races that can eventually break down the resistance.
Project Summary
Overview
Cultivar resistance and crop rotation are the key strategies for management of blackleg on canola. Since the main pathogen that causes blackleg disease in canola in Canada (Leptosphaeria maculans) has a strong ability to evolve and adapt, resistance (R) genes within canola cultivars can be overcome after repeated uses.
As well, at least one virulent race of L. maculans that already exists for most of the R genes available, no new mutation is required and virulent races are present in the current pathogen population. So it is essential to understand the race composition and dynamics in the pathogen population for effective deployment of R genes.
This project analyzes annual L. maculans population on the Prairies (between 2017-2021) provides an important guidance to the selection of effective R genes in blackleg resistance breeding, as well as to the recommendation of canola cultivars carrying proper/effective R genes.
Purpose
This study is part of the continued efforts to provide industry and producers with up-to-date pictures of L. maculans race profile, which can be used to guide the deployment or rotation of canola cultivars carrying different R genes. It can also gain important insights into pathogen race changes in response to resistant cultivars used over the years, allowing industry to be proactive before the erosion of specific resistance R genes.
The key objectives were to:
1) Monitor and analyze the pathogen population to gain insights into race dynamics and population virulence.
2) Provide industry and producers with up-to-date pictures on L. maculans population virulence in the prairie provinces to guide the selection and recommendation of effective R genes for blackleg resistance breeding and disease management.
3) Understand the diversity of L. maculans population and identify pathogen races capable of overcoming common R genes in canola cultivars before widespread breakdown of the resistance.
4) Evaluate, develop and adopt markers for efficient L. maculans race monitoring.
Results
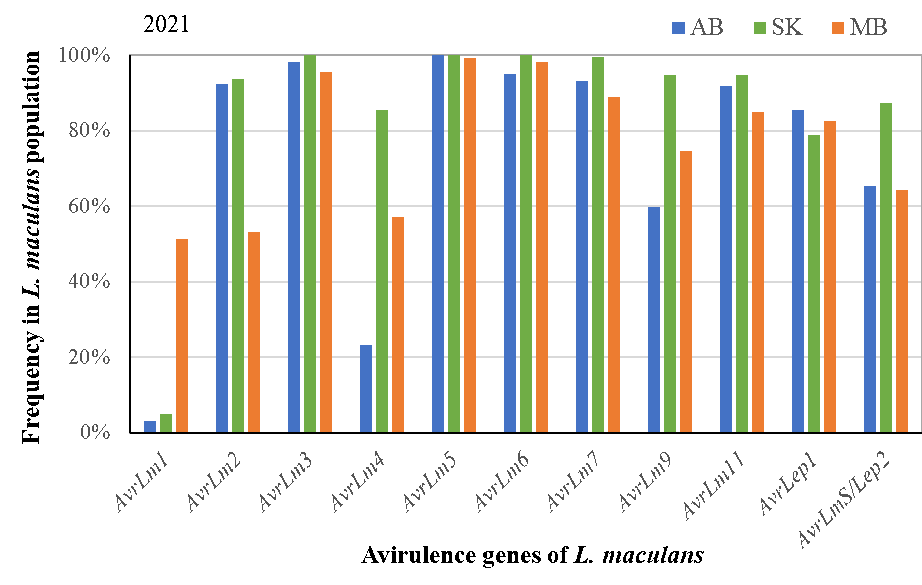
Canola stubble with blackleg symptoms were collected during canola disease surveys organized by the provincial pathologists/coordinators in Alberta, Saskatchewan and Manitoba, respectively. These samples were processed to obtain about 500 L. maculans isolates each year and tested for presence/absence of target Avr genes using KASP markers (AvrLm1,2,3,4,5,6,7,9,11,S-Lep2) and/or host differentials (only AvrLep1 after 2018). In total, about 2,600 L. maculans isolates were analyzed for the entire study over the 5-year period.
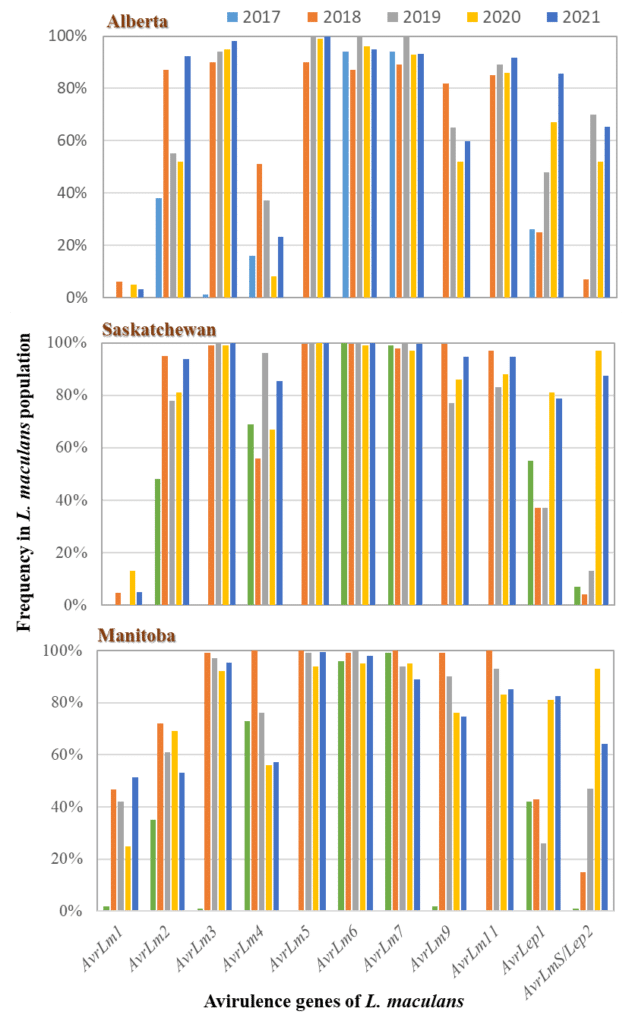
A total of 83 races were identified in the L. maculans population, with top-10 races accounting for 72.3%, 93.7% and 57.1% in Alberta, Saskatchewan and Manitoba. The diversity in pathogen shows the potential for resistance erosion as virulent races had already been present for all R genes, except Rlm10. To manage selection pressure/resistance breakdown, judicious uses of R genes will be important.
The study shows that canola cultivars carrying any of the resistance genes Rlm5, Rlm6, Rlm7, Rlm10, Rlm11 and LepR1 are likely resistant to blackleg in western Canada due to common presence of corresponding Avr genes in the pathogen population. The resistance genes Rlm3 or Rlm9 may be ineffective due to the ‘masking effect’ of AvrLm7 to AvrLm3 and AvrLm9. Rlm4 may be less effective in Alberta due to low levels of AvrLm4.
Conclusions and recommendations
The study identified the pattern of Avr dynamics in the L. population in western Canada between 2017 and 2021, which has prompted the deployment of new R genes, including Rlm2, Rlm4 and Rlm7, in canola cultivars. Using the information, growers and agronomists can better select/rotate canola cultivars with effective R genes for management of blackleg on a regional basis.
As well, judicious deployment of R genes and rotation of resistant cultivars is important for genetic control of blackleg. If an R gene is used repeatedly, there will be selection pressure for virulent races that can eventually break down the resistance.





