Key Result
This project demonstrated that biodiverse agricultural systems improve agricultural productivity, and climate management. The outcomes contribute to a better understanding of canola microbiota, which provide important information to improve canola yields and reduce greenhouse gas emissions by identifying the best rotation system that could potentially reduce nitrogen fertilizer use.
Project Summary
Overview
The objective of this project was to assess the consistency and variability in the composition of the canola core rhizospheric and root microbiota, and determine the crop rotation systems best favouring the establishment of a beneficial root microbiome in canola. The crop rotation systems that enhance the beneficial root microbiome of canola and possibly increase canola productivity while allowing the reduction of fertilizer and pesticide use were also investigated.
Hamel’s research team tested two parameters of this process: the role of soil history in environmental filtering, and the selection pressure created by host plants. They also described the impact of five Brassicaceae plants and their soil history on the structure of bacterial communities in their rhizosphere and roots. It was determined that soil history was significant in structuring the bacterial communities when soil chemistry was highly significant under possible drought conditions. Second, that the Brassicaceae host plants (Brassica napus, B. carinata, B. juncea, Sinapis alba and Camelina sativa) were consistently significant in structuring the bacterial communities.
Summary of key results achieved
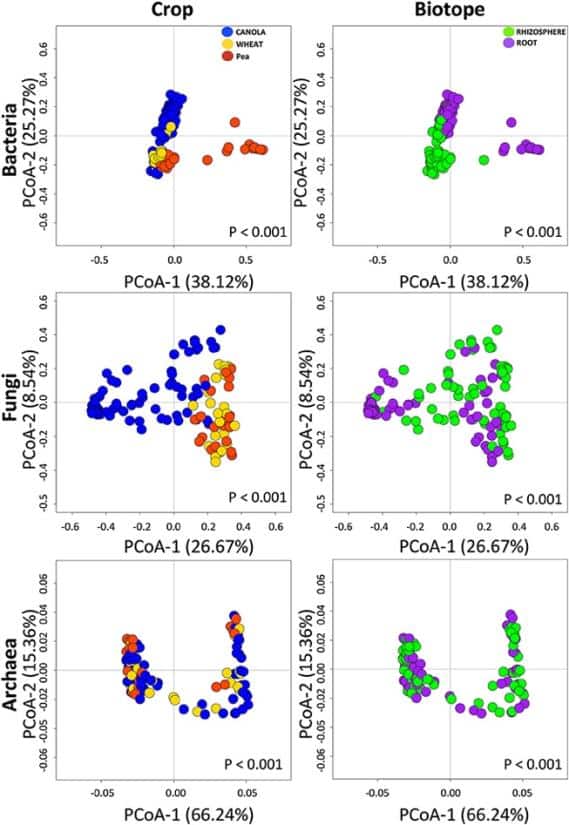
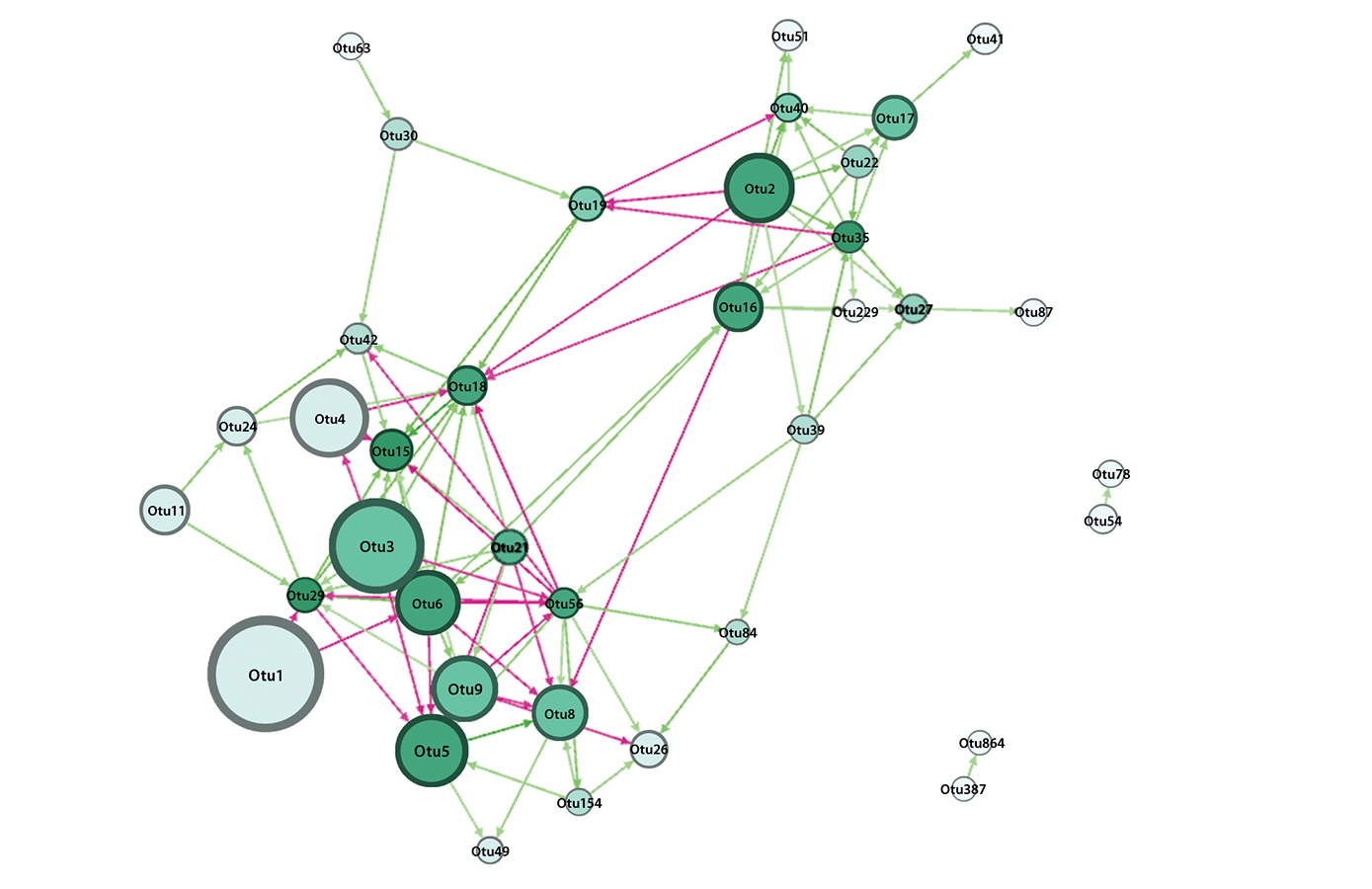
- Canola microbiomes were documented for three communities of microorganisms, namely, bacteria, fungi, and archaea.
- Canola microbiomes were distinguished between the two biotopes (roots and rhizosphere) and were significantly different from those of the reference crops (wheat and pea).
- The research team highlighted the potential PGPR (plant growth-promoting rhizobacteria) among those microorganisms by correlating the core microbiome members in the Canadian Prairies with canola yield.
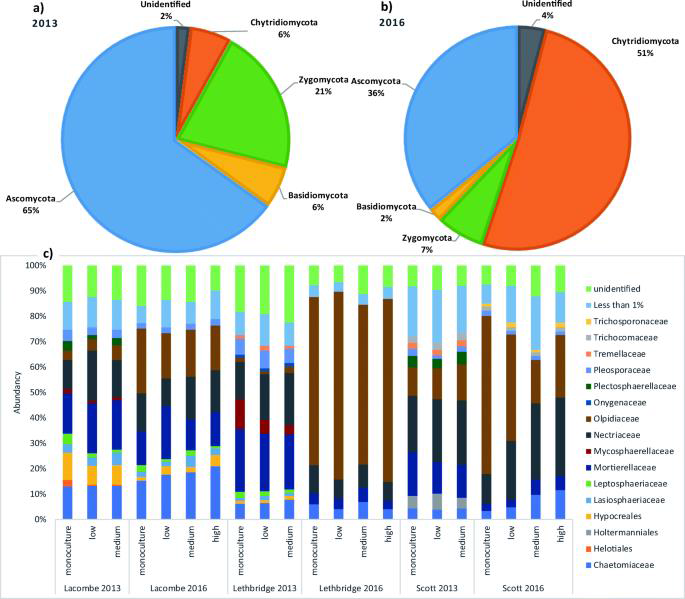
- Fertilization and seeding rates seem to influence certain taxa forming the core and eco microbiomes of canola based on the relative abundances profiles, notably the parasite Olpidium brassicae which was less abundant at the higher seeding rate.
- Certain archaeal taxa showed some specificity to crops and treatments. Furthermore, the putative interactions between the members of bacterial and fungal core microbiomes were weaker with higher fertilization and seeding than the recommended treatments in canola rhizospheres.
- A preceding rotation phase of lentil significantly increased the expression of nifH gene by 23% compared with wheat and improved nxrA gene expression by 51 per cent with chemical fallow in the following oilseed crops respectively.
- Lentil substantially increased biological N2 fixation and reduced denitrification in the following oilseed crops.
- Results revealed that most nitrogen-cycling gene transcripts are more abundant in the microbiomes associated with roots than with the rhizosphere.
- This research brings a new level of understanding on how crop diversification and rotation sequences are related to nitrogen-cycling in annual cropping systems.
- It also provides information about the canola root microbiome that is fundamental for the design of microbiome management strategies for improving canola yield and health.
Conclusions
This project demonstrated that biodiverse agricultural systems improve agricultural productivity, and climate management. The challenge in employing microbiome technologies for improving agricultural systems is the pre-requisite of understanding how soil microbial communities are structured.
The outcomes contributed to a better understanding of canola microbiota which provided important information to improve canola yields and reducing greenhouse gas emission by identifying the best rotation system that could potentially save nitrogen fertilizer uses.
Related publications
This research was reported in these publications:
- Floc’h J-B, Hamel C, Lupwayi N, Harker KN, Hijri M and St-Arnaud M (2020) Bacterial Communities of the Canola Rhizosphere: Network Analysis Reveals a Core Bacterium Shaping Microbial Interactions. Front. Microbiol. 11:1587. doi: 10.3389/fmicb.2020.01587.
- Floc’h, JB., Hamel, C., Laterrière, M. et al. Inter-Kingdom Networks of Canola Microbiome Reveal Bradyrhizobium as Keystone Species and Underline the Importance of Bulk Soil in Microbial Studies to Enhance Canola Production. Microb Ecol (2021). https://doi.org/10.1007/s00248-021-01905-6
- Wang, L., Gan, Y., Bainard, L.D., Hamel, C., St-Arnaud, M. and Hijri, M. (2020), Expression of N-cycling genes of root microbiomes provides insights for sustaining oilseed crop production. Environ Microbiol, 22: 4545-4556. https://doi.org/10.1111/1462-2920.15161





