Key Result
Researchers developed the Canadian Clubroot Differential (CCD) Set, which represents an improved system for the identification of new virulence profiles of Plasmodiophora brassicae and their classification into pathotypes. The CCD Set is an important new tool that can be applied directly on the ground by the sector, including agronomists, breeders and researchers.
Project Summary
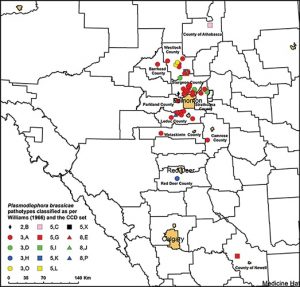
By the end of 2018, more than 3,000 clubroot-infested fields, the majority of which are located in central Alberta, have been confirmed in Western Canada. This represents a large increase since clubroot was first identified in a dozen canola fields in 2003. Although clubroot-resistant (CR) cultivars have been available since 2009, a new strain capable of overcoming resistance to Plasmodiophora brassicae in canola was identified in central Alberta in 2013, which appeared to be highly virulent on all canola cultivars available on the market. The understanding of the new strains of P. brassicae that have emerged in Western Canada in recent years has improved through this study and improved practices for their identification and management have been developed.
The objectives of this three-year project were to monitor the spread of novel clubroot strains through surveys, assess the potential of novel pathotypes to reappear, characterize the pathotypes of clubroot that appear where resistance has broken down and multiply inoculum of novel pathotype(s) for resistance screening. Field surveys were conducted in commercial canola crops in Alberta representing 40 counties and municipalities from 2014 to 2017. The virulence of 151 P. brassicae populations (clubbed roots) was tested on a suite of CR canola cultivars, representing one club from each field in which clubroot was found on a CR canola crop. Surveys identified a total of 17 known pathotypes in Canada, including at least 11 of which can break resistance in CR canola (and five of which are the original “old” ones).
Researchers identified the virulence pattern of each of the P. brassicae populations and assessed the pathotypes on 13 Brassica hosts representing the CCD Set. Each unique virulence pattern on the hosts of the CCD Set was regarded as a distinct pathotype and assigned an identifying letter. The resulting CCD Set has a greater differentiating capacity than the systems that were previously in use, and has allowed the identification of many new pathotypes of P. brassicae that would have otherwise gone undetected. Since this system also includes the differentials of Williams and Somé et al., it allows users to obtain pathotype designations according to those systems as well.

The CCD Set will serve as an effective method to identify novel pathotypes and quickly determine their ability to overcome certain key sources of resistance. The P. brassicae populations representing key pathotypes have been made available to private and public breeders (subject to appropriate biosafety considerations) for screening purposes, in order to assist with the identification of effective resistance sources and the development of new CR canola products. The genomic information obtained and analyses conducted provide an excellent resource for quickly identifying potential molecular markers, and for comparing genetic similarities or differences between different populations of the pathogen. It is clear that an integrated approach, combining other tools in addition to genetic resistance, will be needed for sustainable clubroot control.
Publications that were produced from this research include:
- Holtz, M.D., Hwang, SF. & Strelkov, S.E. Genotyping of Plasmodiophora brassicae reveals the presence of distinct populations. BMC Genomics 19, 254 (2018). https://doi.org/10.1186/s12864-018-4658-1
- Stephen E. Strelkov, Sheau-Fang Hwang, Victor P. Manolii, Tiesen Cao, Rudolph Fredua-Agyeman, Michael W. Harding, Gary Peng, Bruce D. Gossen, Mary Ruth Mcdonald & David Feindel (2018) Virulence and pathotype classification of Plasmodiophora brassicae populations collected from clubroot resistant canola (Brassica napus) in Canada, Canadian Journal of Plant Pathology, 40:2, 284-298, DOI: 10.1080/07060661.2018.1459851
- Michael D. Holtz, Sheau-Fang Hwang, V.P. Manolii & Stephen E. Strelkov (2021) Molecular evaluation of Plasmodiophora brassicae collections for the presence of divergent genetic pathogen populations before and after the release of clubroot resistant canola, Canadian Journal of Plant Pathology, DOI: 10.1080/07060661.2021.1950320





