Key Result
This study identified the prevalence and changes of Avr genes in pathogen populations on the Prairies between 2012 and 2015 which is useful to canola breeding companies for deploying effective resistance genes against the current pathogen population on the Prairies. Fields with severe blackleg damage should consider a longer crop rotation option as the first line of defense. As well, it would be prudent to assess prominent Avr types in the field when using a canola cultivar effective against the pathogen population in a specific field.
Project Summary
Overview
In western Canada, cultivar resistance and crop rotation were the key strategies for successful management of blackleg of canola during 1990s and 2000s. The disease has increased in both prevalence and severity in recent years. Pathogen population shift and tighter crop rotations were suspected to be the primary cause of disease increase. In 2012, a large number of fields with resistant (R) or moderately resistant (MR) cultivars were found with severe blackleg, especially in southern Manitoba and parts of Alberta. Changes in pathogen may produce virulent races that can overcome the R genes in canola cultivars, resulting in resistance breakdown.
To deploy R genes effectively, the pathogen race composition (Avirulence profile or Avr profile) and dynamics should be determined and monitored. This work was initiated in 2007 using nine “Westar” trap plots and found that Avrlm1 and AvrLepR2 were generally low in the pathogen population while the other Avr genes were at moderate to high levels. Another study using samples collected from commercial fields in 2010 and 2011 found that AvrLm3 and AvrLm9 had decreased substantially from the levels observed previously. At the same time, AvrLm7 increased. It was not clear whether the change was caused by different sampling methods used in two studies; theoretically, the pathogen population in trap plots should reflect that in surroundings, but it’s unclear whether the R genes used in commercial fields can skew the data because most these canola varieties carry Rlm3.
The objectives of this project were to analyze/monitor the Avr profile using Westar trap plots for regional pictures of Avr frequencies in the pathogen population. The study can also help identify new pathogen races capable of overcoming specific R genes and the information can help the judicious deployment of specific R genes in canola cultivars for effective control of blackleg.
Methods and results
Between 2012 and 2015, up to 35 Westar trap plots were seeded each year in all major canola production areas on the Prairies. At early crop maturity, diseased stems were sampled from each plot, pathogen isolated and tested on 14 differential brassica varieties/lines carrying known R genes to determine the presence/absence of specific Avr genes in pathogen; plants carrying an R gene would resist the infection only when the pathogen isolate carried the corresponding Avr gene. The number of pathogen isolates tested each year varied slightly depending on the blackleg occurrence in different regions; in a light disease year, fewer isolates were obtained due to a lower rate of isolation, but overall the number ranged from 202 to 626 isolates each year.
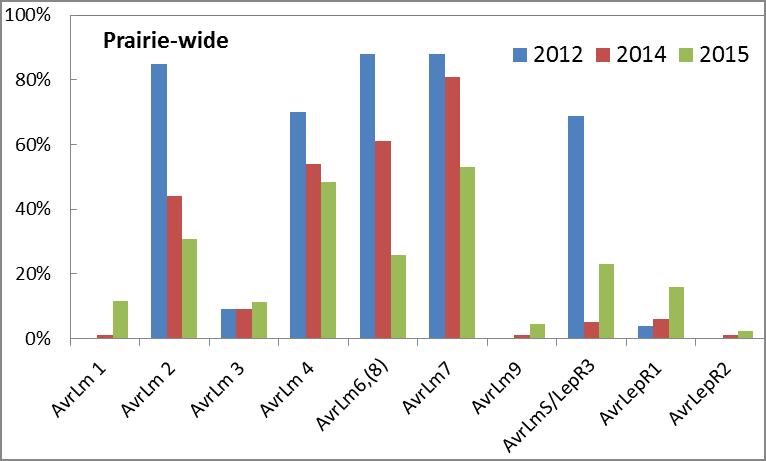
Looking across the Prairies, the pathogen population changed noticeably over the four years of study; AvrLm2, AvrLm4, AvrLm6,(8) and AvrLm7 were present at relatively high frequencies while the other Avr genes were low or even undetectable. AvrLmS was detected in 70% of the isolates in 2012 but was found in only 5% and 23% of the isolates collected in 2014 and 2015, respectively. Even those high-frequency Avr genes observed in 2012 were noticeably lower in 2015. Continued monitoring is warranted to determine if this is a new trend and where the further changes are going. When compare the data from Westar trap plots with those of commercial fields (2015) based on canola disease surveys, the research team noticed that the prevalent Avr genes were similar between the respective populations; AvrLm1, AvrLm3, AvrLm9 and AvrLepR2 were generally very low while AvrLm2, AvrLm4, AvrLm6,(8) and AvrLm7 were high in the field population. For the data in 2015, AvrLm6,(8) appeared noticeably more common in commercial fields than in Westar trap plots.
Among the three provinces, similarities as well as differences were observed in Avr profile; AvrLm2, AvrLm4, AvrLm6,(8) and AvrLm7 were generally at high frequencies in each province while the other Avr genes were very low. AvrLm2, however, seemed to decrease substantially in Alberta (57% to 8%) and Manitoba (36% to 6%) between 2014 and 2015, but held steadily at about 40% in Saskatchewan. In Alberta, the frequency of AvrLm4 was consistently lower (∼40%) than those in Saskatchewan and Manitoba (60-80%) over the years of study.
Conclusions and recommendations
This study identified the prevalence and changes of Avr genes in pathogen populations on the Prairies between 2012 and 2015; AvrLm2, AvrLm4, AvrLm6,(8) and AvrLm7 were found at high frequencies while AvrLm1 and AvrLm3 were generally low. This information is useful to canola breeding companies for deploying effective R genes against the current pathogen population on the Prairies.
Since only the Rlm1 and/or Rlm3 were found commonly in Canadian cultivars, it is likely that the major-gene resistance to blackleg is currently lacking on the Prairies due to the generally low presence of AvrLm1 and AvrLm3 in the pathogen population. Therefore, it is postulated that the blackleg resistance by many of the R-rated canola cultivars is via other mechanisms, especially race nonspecific resistance, or quantitative/adult plant resistance. It is expected that Rlm4, Rlm6 or Rlm7 can substantially improve the blackleg resistance in most regions on the Prairies, especially when used with proven background resistance.
Caution should be exercised when using this big-picture information to guide canola cultivar rotation based on specific R genes in a given field because the pathogen race structure can change substantially from one field to another as shown by the results of study. In the cases where severe blackleg damage occurs in a field, it would be a good practice to consider a longer crop rotation option as the first line of defense. To use a canola cultivar effective against the pathogen population in a specific field, it would be prudent to assess prominent Avr types in the field.





