Key Result
Clubroot continues to spread, but improved monitoring and methodologies allow for more pathotype characterization to better identify, quantify and eventually manage clubroot-causing pathotypes going forward.
Project Summary
Overview
As the appearance of new clubroot pathotypes capable of overcoming resistance is one of the most important threats facing canola production in clubroot-infested regions of the Prairies, this research led by Steve Strelkov focused on tracking and responding to changes in the clubroot pathogen.
Surveillance in Alberta: In addition to clubroot being found in 79 of the 543 canola crops surveyed in 2018, another 221 cases were found by municipal personnel.
These cases are distributed across 40 counties and municipal districts, plus Edmonton, Medicine Hat and the town of Stettler. Disease identification in new locations suggested that clubroot is spreading in the Peace Country of northwestern Alberta and in southern Alberta.
In most cases, clubroot severity was mild or moderate, although five crops were heavily infested – two of which had included clubroot-resistant canola cultivars – and
pathogen populations from these fields were collected for further evaluation in the lab.
Surveillance in Saskatchewan: Of the 1,800 fields surveyed, 37 canola crops were found with visible clubroot symptoms. A subset of 10 samples was sent for pathotype characterization and nine samples were sent for pathotyping.
Surveillance in Manitoba: Of the 100+ fields in 2018, 17 cases of clubroot were identified, bringing the cumulative total to 33 confirmed infestations since 2009.
A subset of six clubroot samples was sent for pathotype characterization, while three collections were sent for pathotyping.
Characterization of single-spore and field isolates:
Thirteen pathotypes were identified amongst the 39 single-spore isolates (populations) obtained and characterized for their virulence on a suite of CR canola cultivars and on the Canadian Clubroot Differential Set.
A total of 119 Plasmodiophora brassicae populations, including 103 from Alberta, 10 from Saskatchewan and six from Manitoba were tested. Eighty-seven of the 103 field isolates collected from Alberta could overcome clubroot resistance, bringing the total number of confirmed fields with resistance issues to 191 in the province.
Of the 10 samples from Saskatchewan and the six field isolates from Manitoba tested, only one isolate from Manitoba could overcome clubroot resistance in CR canola, while the rest did not. This was the first confirmed case of a clubroot resistance-breaking pathotype detected on canola outside of Alberta.
Refinement of the clubroot differential set: Collectively, the results identified 19 Brassica accessions that had good ‘differential capability’ and potential as differential
hosts. These included two B. napus, eight B. rapa and nine B. nigra accessions.
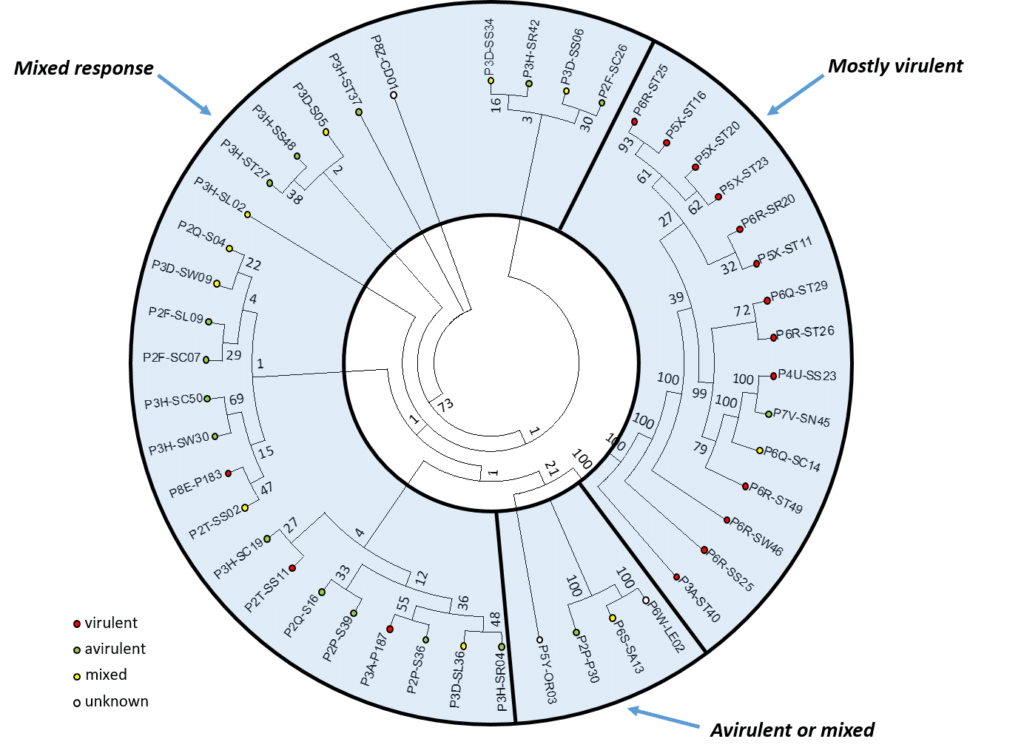
Examination of the genetic and virulence relationships between pathotypes and their interactions: Resting spores representing 45 isolates of P. brassicae, including the 39 single-spore isolates described earlier in this report plus an additional six field isolates, were selected for genomic analysis. This allowed for the development of a binary matrix to construct a dendrogram showing relationship between 45 P. brassicae field and single-spore isolates.
Development of molecular tests to distinguish between pathotypes: Successful analysis involving 45 P. brassicae genomes initiated the development of molecular markers for the identification of P. brassicae pathotypes using the genome sequence information (which is highly specific and more accurate than standard PCR). This can be used to identify and quantify clubroot-causing pathotypes from plant, soil and water samples.





