Step 1. Sample Collection
The ideal time to pull and assess plants for blackleg infection is at 60% seed colour change, which is around swath timing. Plants should still be green. Cut just below the crown of the plant into the root material to assess.
“Look for black discolouration within the hypocotyl tissue, often appearing in a wedge pattern. If a stem looks to be infected, keep a 2-3 inch piece for testing. Allow samples to air dry overnight before packaging to avoid excessive fungal growth.” From Discovery Seed Labs’ Blackleg Sampling Factsheet
Note: Old canola residue samples that contain pycnidia (the black spots) can be submitted, but these samples may show many other fungal species that are more saprophytic than L. maculans
Step 2. Lab analysis
The lab will determine which blackleg species is present: L. biglobosa or L. maculans.
L. biglobosa, a mild or weakly virulent species, is often associated with upper stem lesions and is widespread in western Canada. This species usually infects plants late in the season, rarely causing significant yield losses and is therefore considered a minor problem
L. maculans is the virulent species causing blackleg. It infects canola from the seedling stage onward, and progressively damages the plant by girdling stems and restricting moisture and nutrient uptake, leading to yield loss. Determining the species is difficult to complete visually. DNA must be taken to determine species.
Step 3: Interpreting the results
Lab reports typically report both the genotype and phenotype of the blackleg races identified from the samples collected.
Genotype is the genetic content or genes/DNA present in the organism – in this case the blackleg fungus.
Phenotype is the physical expression of genes; it may or may not be influenced by environment.
Producers need to look at the estimated phenotype when making variety selections. The phenotype is estimated because a physical expression test is not completed where different canola lines are grown out and tested with the inoculum collected. The phenotype is estimated based on published literature looking at gene interactions.
Lab reports will identify the predominant race of blackleg or give a frequency breakdown of blackleg races found within the samples collected. Major genes within canola varieties need to match up to avirulence genes (Avr) within the pathogen to be effective.
Lab result, example 1. Phenotype: AvrLm4-5-6-7-11
Lab result, example 2. Phenotype: AvrLm2-4-5-6-7-11 (25%)
AvrLm4-5-6-7-11 (50%)
AvrLm1-4-5-6-7 (16%)
In example 2, you want to find a major gene that corresponds with an avirulence gene found in all the races listed. If a diversity of blackleg races are presented, find a major gene that matches to the most frequent race. In this example, avirulence gene 4 is found in all blackleg races identified. Using a variety deploying a major gene of Rlm4 (or Gr E1) would be a suitable match.
Step 4: How to use the results
Here are the Resistance Group (RG) labels and the corresponding major genes in Canadian canola cultivars:
RG A = Rlm1 or LepR3*
RG B = Rlm2
RG C = Rlm3
RG D = LepR1
RG E1 = Rlm4**
RG E2 = Rlm7**
RG F = Rlm9
RG G = RlmS
RG H = LepR2
RG X = unknown
Reference blackleg.ca section on R-gene rotation
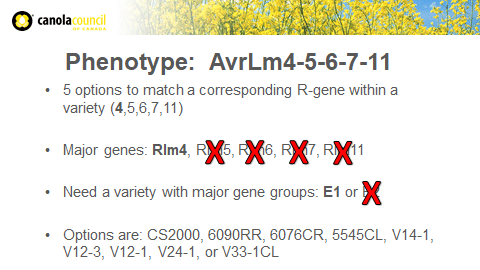
Example Scenario:A producer grew a canola variety from resistance group C and saw severe disease symptoms. So the producer submitted samples to a lab. The lab results showed Genotype AvrLm3-4-5-6-7-9-11 and Phenotype AvrLm4-5-6-7-11 present in the samples.
Phenotype results show five options to match a corresponding R-gene within a variety. The Avr genes are 4, 5, 6, 7 and 11, which means the producer can choose canola varieties with one of these major genes: Rlm4, Rlm5, Rlm6, Rlm7, Rlm11.
Currently, the only major gene available on this list is Rlm4 (from resistance group E1). Variety options with Rlm4 or resistance group E1: CS2000, 6090RR, 6076CR, 5545CL, V14-1, V12-3, V12-1, V24-1, or V33-1CL. If the seed this producer wants does not provide this option, then the producer should still switch varieties and look to grow a strong Quantitative resistant variety.
The producer could take a risk and continue growing group-C varieties, but if the environmental conditions are conducive to disease development, the crop is at risk for infection as the major gene being deployed (in this example Rlm 3) does not match up to the predominant blackleg race in the field and therefore is not effective.
More resources and further reading:
Discovery Seed Labs blackleg test
Manitoba Pest Surveillance Lab blackleg sampling guidelines
Canola Encyclopedia chapter on blackleg
Gary Peng, AAFC research scientist, webinar on blackleg